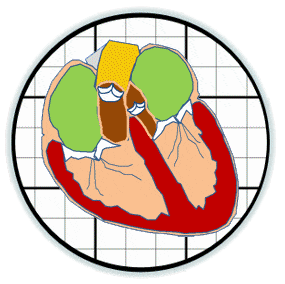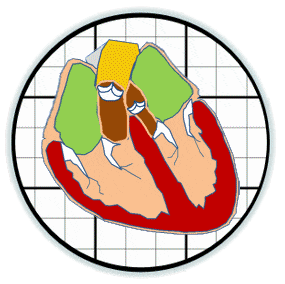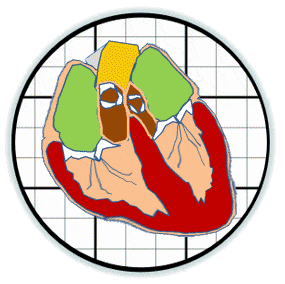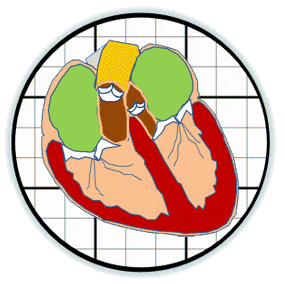
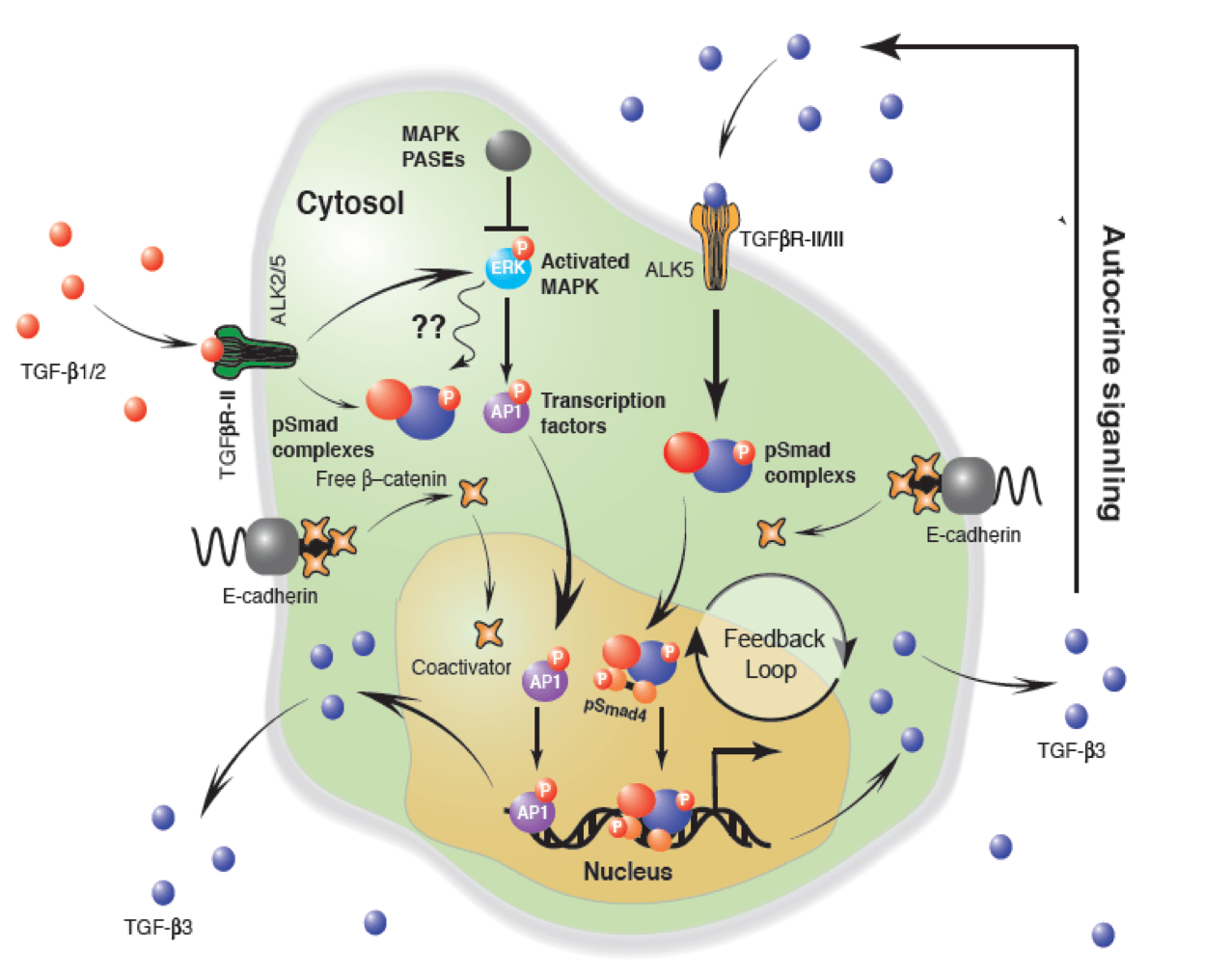
 Heart valves initiate as specific regions of endocardium within the atrioventricular canal and outflow tract of the heart tube that undergo an endocardial to mesenchymal transformation (EMT), forming valvuloseptal primordia called cushions. The molecular regulation of cushion formation is well studied; however, defects in these early events are not survivable. Much less is known about the events regulating Post-EMT cushion remodeling into valves. This involves three simultaneous processes: (1) elongation and condensation of the tissue into thin leaflets or cusps, (2) mesenchymal differentiation to a fibroblastic phenotype, and (3) acquisition of a stratified extracellular matrix (ECM) architecture. Defects in these later remodeling events are of strong clinical concern because they are survivable but immediately life-threatening or predispose the valve to premature structural failure. While regulatory molecules have been identified, including ligands (e.g. TGF–β, BMP, VEGF, Jag/Dll, Wnt, EGF) and their receptors, transcription factors (e.g. smads, snail/slug, Notch, Tbx5, Twist1, Tbx20, Sox9, Scleraxis, NFATc1, β-catenin), and matricellular proteins (e.g., hyaluronic acid, versican, periostin), the important coordination of these pathways remain unclear. Genetic deletion of many of these individual genes result in similar malformation outcomes that are mechanically insufficient. Hemodynamic maldistribution has been postulated as a causal mechanism of clinical CHD, but before our work was almost always studied as a consequence of genetic/biological perturbations rather than the other way around. We test novel hypotheses that these molecular interactions underlie coordinated cellular decisions in response to their hemodynamic environment to correctly elongate and remodel the valvuloseptal apparatus. Elucidation of these multi-scale bio-engineering programs is a critical need for bio-inspired intervention strategies to improve morphological and functional outcomes.
Heart valves initiate as specific regions of endocardium within the atrioventricular canal and outflow tract of the heart tube that undergo an endocardial to mesenchymal transformation (EMT), forming valvuloseptal primordia called cushions. The molecular regulation of cushion formation is well studied; however, defects in these early events are not survivable. Much less is known about the events regulating Post-EMT cushion remodeling into valves. This involves three simultaneous processes: (1) elongation and condensation of the tissue into thin leaflets or cusps, (2) mesenchymal differentiation to a fibroblastic phenotype, and (3) acquisition of a stratified extracellular matrix (ECM) architecture. Defects in these later remodeling events are of strong clinical concern because they are survivable but immediately life-threatening or predispose the valve to premature structural failure. While regulatory molecules have been identified, including ligands (e.g. TGF–β, BMP, VEGF, Jag/Dll, Wnt, EGF) and their receptors, transcription factors (e.g. smads, snail/slug, Notch, Tbx5, Twist1, Tbx20, Sox9, Scleraxis, NFATc1, β-catenin), and matricellular proteins (e.g., hyaluronic acid, versican, periostin), the important coordination of these pathways remain unclear. Genetic deletion of many of these individual genes result in similar malformation outcomes that are mechanically insufficient. Hemodynamic maldistribution has been postulated as a causal mechanism of clinical CHD, but before our work was almost always studied as a consequence of genetic/biological perturbations rather than the other way around. We test novel hypotheses that these molecular interactions underlie coordinated cellular decisions in response to their hemodynamic environment to correctly elongate and remodel the valvuloseptal apparatus. Elucidation of these multi-scale bio-engineering programs is a critical need for bio-inspired intervention strategies to improve morphological and functional outcomes.
Selected Projects:
Unraveling the Role of Mechanical Stimulation on Cardiac Developmental Maturation
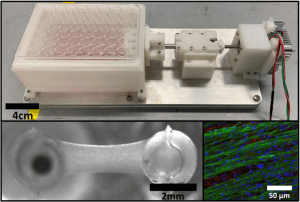 There remains a significant clinical need to develop cardiac regenerative therapies to restore heart function following a myocardial infarction. Engineering cardiac tissues as a living tissue replacement remains a promising approach. However, due to the lack of maturity of these engineered cardiac tissues, they often fail to integrate with the host tissue, have poor survivability, and lead to unwanted arrythmias upon implantation in vivo. One such way to improve engineered cardiac tissue maturity is through the use mechanical stimulation, however, current systems utilize uninspired non-physiological conditioning regimens. Therefore, we have developed a novel customized in vitro engineered cardiac tissue platform and high throughput mechanical stimulation bioreactor system that can recapitulate the cardiogenic stretch patterns of natural development. Utilizing this system, we are investigating how specific mechanical stimulation regimens affect developmentally staged embryonic chick cardiac cell maturation in a spatiotemporal manner. We also are developing various quantitative image analysis tools to better ascertain how specific mechanical forces affect the spatial organization and functionality of these cardiac cells.
There remains a significant clinical need to develop cardiac regenerative therapies to restore heart function following a myocardial infarction. Engineering cardiac tissues as a living tissue replacement remains a promising approach. However, due to the lack of maturity of these engineered cardiac tissues, they often fail to integrate with the host tissue, have poor survivability, and lead to unwanted arrythmias upon implantation in vivo. One such way to improve engineered cardiac tissue maturity is through the use mechanical stimulation, however, current systems utilize uninspired non-physiological conditioning regimens. Therefore, we have developed a novel customized in vitro engineered cardiac tissue platform and high throughput mechanical stimulation bioreactor system that can recapitulate the cardiogenic stretch patterns of natural development. Utilizing this system, we are investigating how specific mechanical stimulation regimens affect developmentally staged embryonic chick cardiac cell maturation in a spatiotemporal manner. We also are developing various quantitative image analysis tools to better ascertain how specific mechanical forces affect the spatial organization and functionality of these cardiac cells.
Mechanobiology of Fetal Valve Remodeling
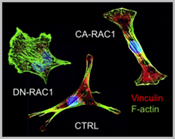
We study how mechanical forces control cell polarity and heterogeneous collective cell migration that we believe is essential for coordinated valve remodeling by related cellular fields (e.g. neural crest, anterior heart field, and epicardium). We hypothesize this is achieved through the coordination of integrin and cadherin signaling. We further utilize our bioreactor technologies to assess the mechanobiological and biomechanical influences integrin/cadherin signaling in valve formation/remodeling events. Among the key downstream mediators of cellular interaction with their environment (e.g. migration, contraction, Figure 1) are small GTPases such as RhoA and Rac1. We further identify novel interacting partners at these sub-cellular interfaces and elucidate their role using molecular gain/loss of function assays. At the cellular scale, we identify endocardial and mesenchymal specific requirements of adhesive signaling to coordinate tissue remodeling ECM patterning in vitro and in vivo. We study these in vivo using both avian and mouse model systems, the former with the benefit of our unique mechanical perturbation techniques, and the latter using cell type specific Cre drivers. We further test how these mechanically regulated adhesive signaling programs orchestrate known valvulogenic signaling pathways, and identify novel regulators via unbiased high throughput nucleotide sequencing (e.g. mRNA, miRNA). We then determine whether these same signaling mechanisms and pathways are involved in postnatal valve homeostasis and if their disruption regulates myxomatous or calcific degeneration using our previously published approaches. With this multi-scale mechanistic insight, we then test whether rebalancing adhesive signaling (first genetically and then via small molecule or targeted nanoparticles) is sufficient to improve fetal morphogenetic outcomes and/or inhibit postnatal valve degeneration.
Hemodynamic Regulation of Valve and Great Vessel Morphogenesis
Only about 10% of human CHD cases have an identifiable genetic cause. Causal interpretations of genetic deletions (even lineage restricted) are confounded by variable penetrance and genetic compensation as development progresses. We utilize our innovative local hemodynamic gain/loss experimental and computational strategies to directly elucidate the molecular, cellular, and morphogenic adaptations that are orchestrated by hemodynamic forces, both to promote normal and malformed outflow valves and great vessels. We can create focal ablations or obstructions within the beating embryonic heart, the proximal/distal outflow tract, and the pharyngeal arch artery network (which collectively form the dual outflow valves and great vessels) in live avian embryos using our innovative femtosecond laser ablation technology. We directly quantify the early and late in vivo consequences of these focal lesions using high frequency ultrasound, in vivo micro-CT, and CFD simulations. We determine how these hemodynamic perturbations affect local cell lineage migrations (via established quail-chick chimeras and/or local electroporation based labeling and genetic modification, cell phenotypes and matrix composition. Armed with this fundamental knowledge, we explore how in vivo ablation of hyperplastic cushions and/or local tissue obstructions, or directed hemodynamic redistribution through local vascular occlusion of can counteract genetic mutations in cardiac and great vessel morphogenesis.
Dynamic Quantitative Imaging of Live Embryogenesis via Micro-CT
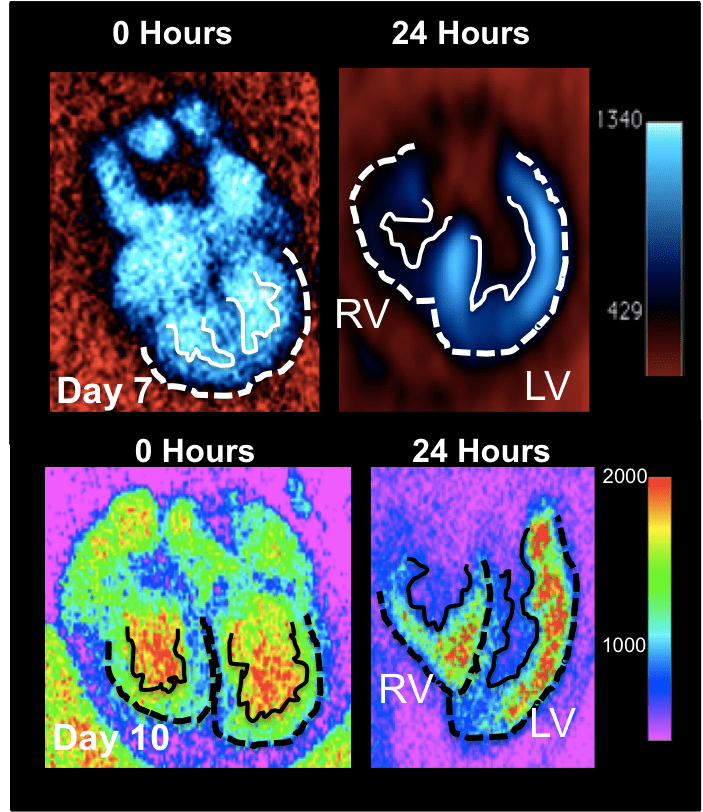
The vast majority of our understanding of multi-scale ventricular remodeling has been inferred via fixed specimens, precluding elucidation of how evolving compact and trabeculated myocardial architecture controls dynamic cardiac output and downstream morphogenesis and defect propagation. Quantitative live imaging technology is essential to unlock these secrets, but most embryonic imaging studies have focused on the earliest heart formation events using optical imaging techniques. Recent studies using optically advantageous zebrafish have highlighted mechanistic connections between forces and ventricular remodeling, but this model cannot answer clinically relevant questions involving septation, left and right sided ventricular myocardial remodeling, and its relationship to valve and outflow vessel morphogenesis and alignment. The avian embryo is a well-established experimentally and genetically accessible model system to answer these questions. Other technologies like high frequency ultrasound or magnetic resonance lack the spatial resolution, contrast, and/or 4D capacity for analyses of live embryonic ventricular architecture analysis. Micro-computed tomography (microCT) is an attractive modality to noninvasively image the avian embryo in 3D with the high spatial and temporal resolution needed to capture the dense and tortuous anatomical changes longitudinally throughout the full range of cardiac morphogenesis. We are establishing and validating the requisite but currently unavailable technology to enable dynamic volumetric imaging of intra-beat and longitudinal ventricular morphogenesis and function. These include: novel exogenous contrast media, prospective gating technology, and quantitative algorithms. We are developing size specific nanoparticles that exhibit unique CT contrast persistence and volumetric biodistribution characteristics during avian embryonic cardiogenesis. We have further developed novel strategies to quantitatively map molecular and cellular expression in 3D using CT contrast. We also develop and utilize novel gating and reconstruction algorithms to quantify the dynamic and multi-scale 5D (3D in space + 2D in time) ventricular, valve, and vascular wall architecture changes across the cardiac cycle and at different stages. We use these technologies to test how targeted alteration of cardiac formation and/or function via genetic or mechanical alterations controls downstream remodeling events in the same embryo over time, integrating hemodynamic, morphological, structural, and molecular datasets.
Multi-scale Computational Systems Mechanobiology of Valve Morphogenesis and Remodeling
We continue to build on our computational innovations to both quantify and predict local and global cardiac geometry changes resulting from hemodynamic and/or genetic changes. We advance computational modeling strategies that accommodate growth within fluid and tissue mechanical fields. This requires unique fluid-structure interaction, constitutive theory, and geometry/mesh adaptation innovations. We further add to our discrete models biological elements such as cells, soluble chemicals, and extracellular matrix proteins in a grid of cellular nodes that can proliferate, move, and/or apoptose. Each cell also contains a molecular systems network that incorporates known signaling pathways that will define how it responds to external molecular and mechanical stimuli. We will predict cross-pathway bottlenecks in valvulogenic signaling that are intractable experimentally or through sequencing data alone. We utilize statistical distribution models to reduce parameter variables to their minimum essential sets, while preserving our capacity for prediction of emergent phenomena. We can directly test these predictions via our unique in vitro and in vivo experimental systems. These approaches enable us to identify master regulators of development that act across multiple signaling networks and biological scales.